DNA Replication Concept Map
Have you ever wondered why offspring often resemble their parents? Why do Black parents not give birth to a white child? These thoughts emerge in the minds of students who observe the variations in the human race around them.
If these questions also strike your mind, know that it is the game of DNA replication. DNA replication is a vital process for passing genetic material from parents to offspring. It ensures that each new generation receives a complete set of genetic instructions from its parents. Hence, the child gets the traits of his parents.
In this blog, we will explain the complete DNA replication concept map to help you understand how genetic material is transferred from parents to offspring.
What is DNA Replication
DNA replication is the process by which the DNA of a genome is exactly copied in cells. In this process, two identical copies of DNA are produced from the original molecule. It is important for cell growth, repair, and reproduction in organisms.
DNA replication transmits genetic information. Without accurate DNA replication, offspring would not receive the correct genetic blueprint from their parents. Due to this reason, continuity of life and the inheritance of traits would not be possible.
DNA replication is usually asked in medical college entry tests. If you are preparing for the MCAT, get help here.
Understanding the Structure of DNA
Before going through the process of replication, it is important to understand the structure of DNA.
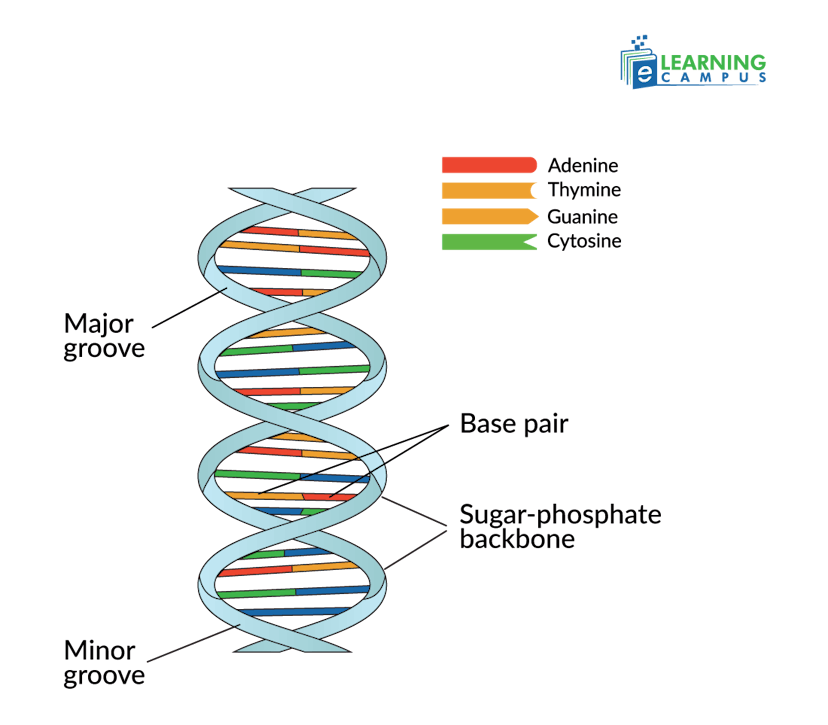
DNA (deoxyribonucleic acid) is a genetic material made up of sugar-phosphate with nitrogen bases. It is a double helix, shaped like a twisted ladder, with two strands that are composed of alternating sugar-phosphate backbones.
- Nucleotides: Each strand of DNA is a polymer of nucleotides.
- Sugar-Phosphate Backbone: Each nucleotide consists of a deoxyribose sugar and a phosphate group. It forms the sturdy backbone of the DNA strand.
- Nitrogen Bases: Attached to the sugar is one of four nitrogenous bases, such as adenine (A), cytosine (C), guanine (G), or thymine (T).
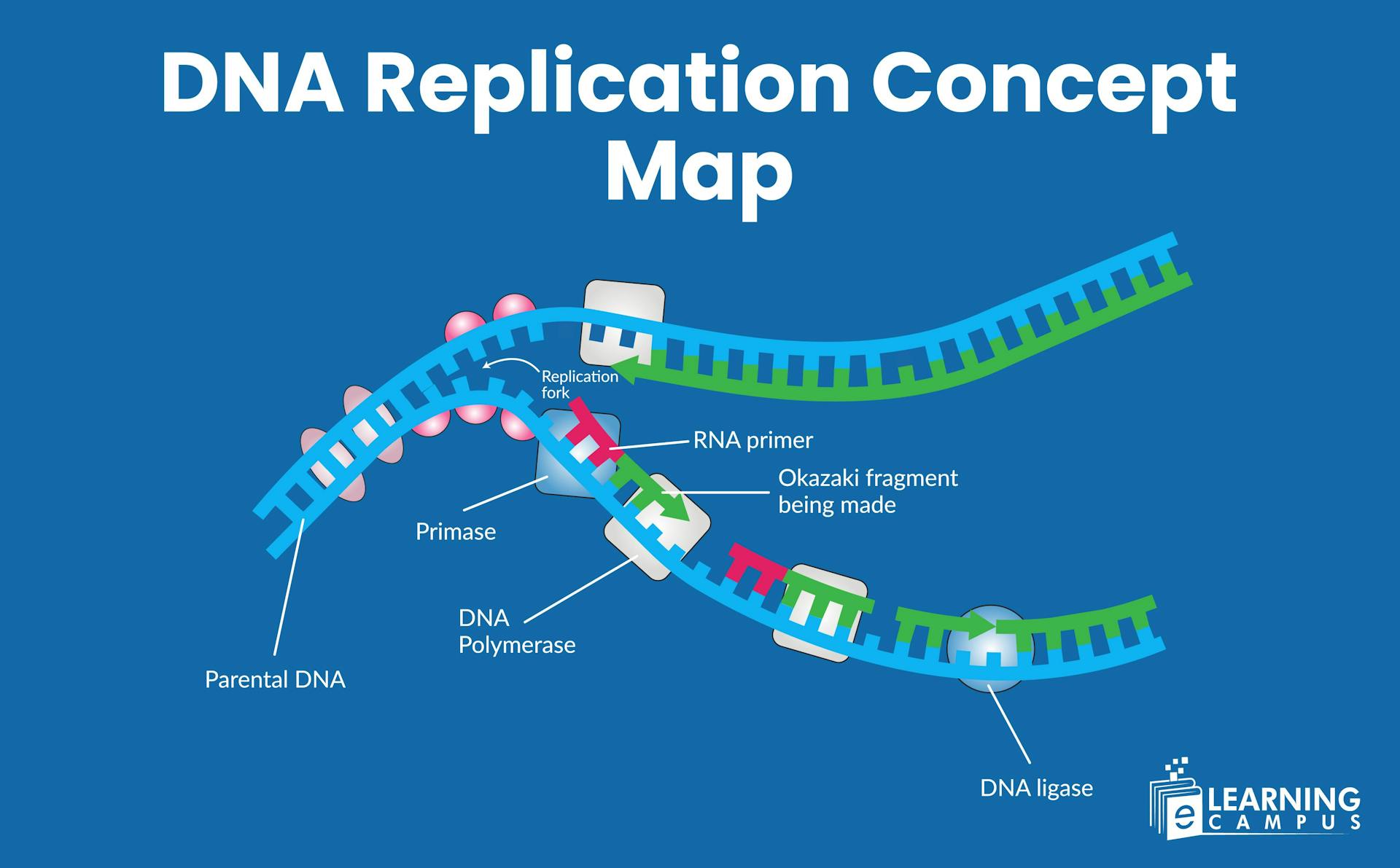
Steps and Process of DNA Replication- Concept Map
The DNA replication process includes three main stages. They are initiation, elongation, and termination. Each of these steps plays an important role in the replication process and transmission of genetic material. You can understand the concept through a DNA replication diagram.
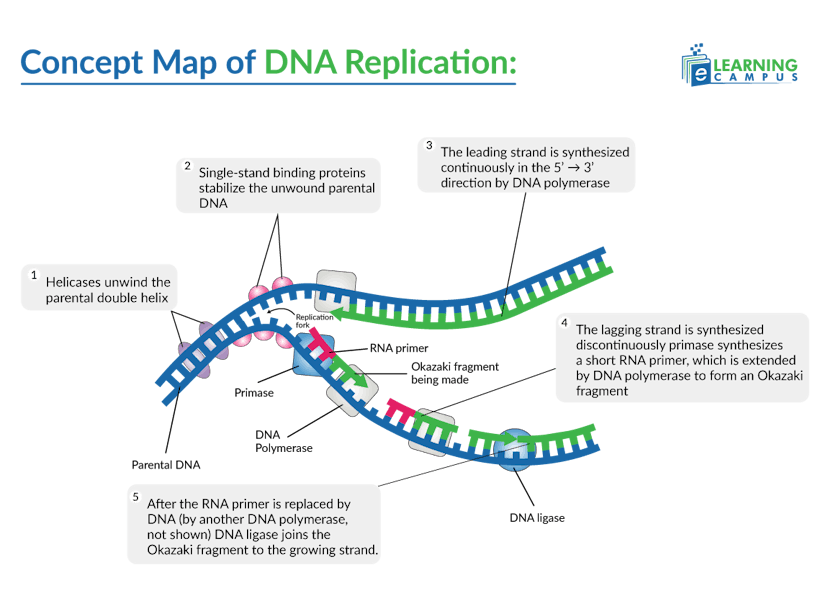
Initiation
Initiation is the initial process of DNA replication. It begins when initiator proteins bind to origins of replication on the DNA, recruiting the helicase enzyme. The initiation involves the following process.
Origin Recognition
The process starts at specific DNA sequences called origins of replication. Initiator proteins, such as the Origin Recognition Complex (ORC) in eukaryotes, recognize and bind to it.
Unwinding
Enzymes like helicase bind to the origin and unwind the double helix by breaking the hydrogen bonds between the base pairs. It creates a Y-shaped structure called the replication fork.
Stabilization of Single Strands
Single-stranded DNA-binding proteins (SSBs) bind to the separated strands of DNA. It prevents them from re-annealing and protects them from degradation.
Primer Binding
DNA replication starts when a short RNA piece called a primer attaches to the DNA strand. The primer acts like a starting signal that tells the cell, ‘begin copying here.’ An enzyme called DNA primase makes these primers and places them where replication should begin.
Elongation
Elongation is the second phase of DNA replication. In this process, Enzymes known as DNA polymerases create the new strand of DNA. It involves the following process.
Primer Extension
DNA polymerase binds to the RNA primer, which is synthesized by primase, and starts adding DNA nucleotides.
Continuous Synthesis (Leading Strand)
On the leading strand, DNA polymerase moves in the 5' to 3' direction towards the replication fork, continuously adding nucleotides as the DNA unwinds.
Discontinuous Synthesis (Lagging Strand)
On the lagging strand, which runs in the opposite direction of the replication fork, DNA polymerase synthesizes DNA in short fragments. This process requires multiple RNA primers and results in the formation of Okazaki fragments.
Nucleotide Addition
For each nucleotide added, the DNA polymerase uses the template strand as a guide, selecting the complementary base (A pairs with T, and C pairs with G).
Primer Removal
After DNA fragments are synthesized, enzymes like DNA polymerase I (in bacteria) or other proteins (in eukaryotes) remove the RNA primers.
Fragment Joining
DNA ligase then seals the remaining gaps between the DNA fragments, creating a continuous DNA strand.
Termination
Termination is the last step in the DNA replication process. In this process, the two replicating forks meet, leading to the complete unwinding and replication of the remaining DNA. The termination process involves the following steps.
Replication Fork Convergence
Two replication forks moving in opposite directions meet. It leads to the full unwinding of the last stretch of parental DNA between them.
Gap Filling and Ligation
Any remaining gaps in the newly synthesized daughter strands are filled in. The nascent strands are ligated (joined together) to create continuous DNA molecules.
Catenane Removal
Double-stranded DNA intertwinings, called catenanes, are resolved and removed, which is crucial for separating the two newly formed circular DNA molecules in bacteria.
Replisome Disassembly
The replication machinery, known as the replisome, is disassembled, and the replication proteins are unloaded from the DNA.
DNA Replication Enzymes and Their Functions
Enzymes play an important role in DNA Replication. They accurately copy genetic information and ensure that it is correctly transferred to new cells. They prevent genetic disorders. The key enzymes that take part in DNA replication are;
Helicase
The Helicase enzyme unwinds and unzips the DNA double helix by breaking the hydrogen bonds between the bases. Due to this breakdown, it creates a replication fork where new strands can be synthesized.
Topoisomerase
This enzyme relieves the torsional strain and prevents the DNA from becoming tangled when helicase unwinds it. It allows replication to proceed smoothly.
Primase
This enzyme synthesizes short RNA primers that provide a starting point with a free 3'-hydroxyl group. It is required by DNA polymerase to initiate the addition of new nucleotides.
DNA Polymerase
The polymerase is the main synthesis enzyme. It adds complementary nucleotides to the 3' end of the new DNA strand, following the base pairing rules. It also has a crucial proofreading function that removes mismatched nucleotides to ensure accuracy and minimize errors.
DNA Ligase
The DNA Ligase connects the newly synthesized DNA segments. It is particularly important in the Okazaki fragments on the lagging strand. It seals the gaps to form a continuous and complete DNA strand.
Single-Strand Binding (SSB) Proteins
These proteins bind to the separated single strands of DNA. They prevent the DNA strands from reannealing (sticking back together) and keep them accessible for replication.
DNA Replication Model
There are three basic DNA replication models. They include the semiconservative, conservative, and dispersive models. These models explain different ways a DNA molecule could be copied, specifically how the original parental strands and newly synthesized daughter strands would be distributed into new DNA molecules after replication.
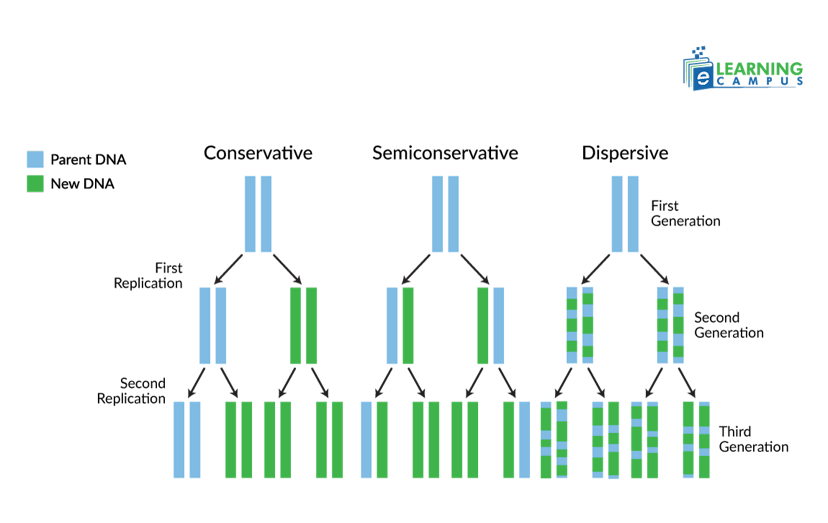
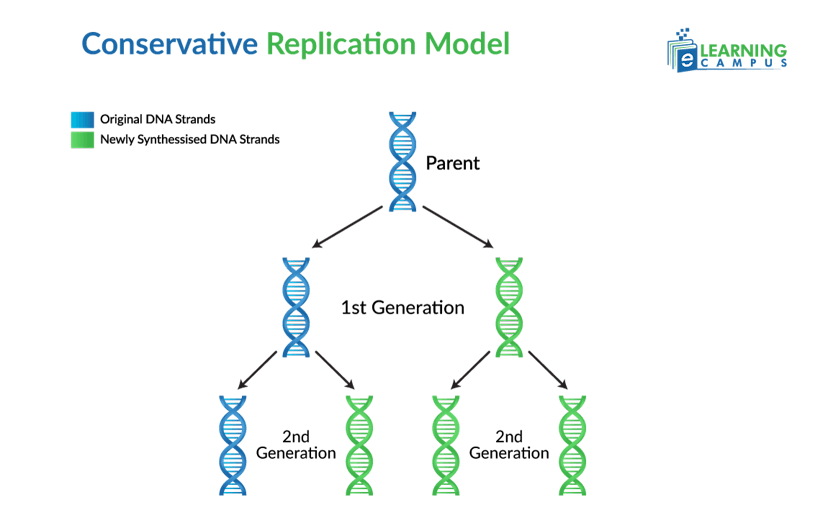
Conservative Model
The conservative model was one of the proposed models of DNA replication. It was later refuted by the Meselson-Stahl experiment. This model states that after DNA replication takes place, the original parental DNA molecule remains intact. The two new daughter molecules each consist of two newly synthesized strands.
It is called conservative because the original DNA molecule remains conserved in this model.
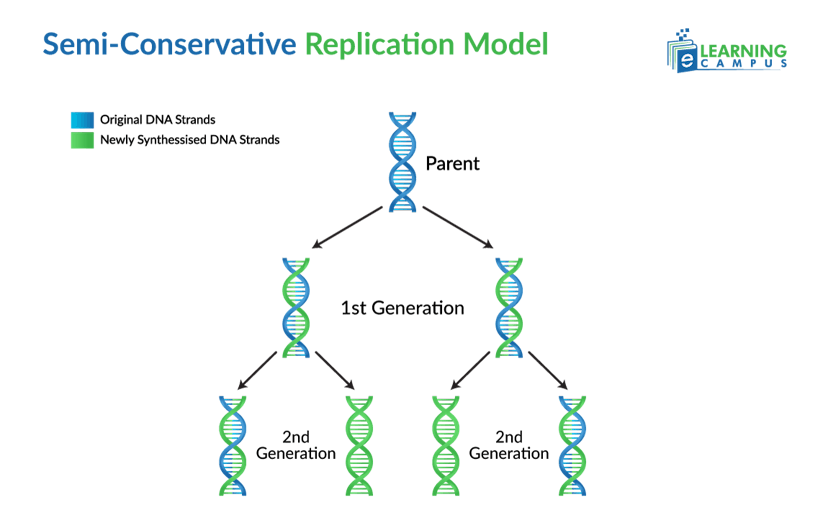
Semiconservative Model
The semiconservative model of DNA replication states that during DNA replication, the double-stranded DNA molecule separates into two single strands. Each original strand serves as a template to synthesize a new complementary strand. The result is two new DNA molecules, each containing one original ‘parental’ strand and one newly synthesized ‘daughter’ strand.
It is called semiconservative because half of the original DNA molecule is conserved in each new molecule.
Dispersive Model
The dispersive model states that the DNA replication results in two DNA molecules. Those molecules are mixtures or hybrids of parental and daughter DNA. It means that the parental DNA breaks into fragments, and new DNA is formed in patches.
Deep learning about DNA replication models with expert science tutor can help you crack the challenging entry exam tests.
DNA Replication in Prokaryotes and Eukaryotes
The core objective of DNA replication is the same in prokaryotes and eukaryotes, but the mechanisms differ due to variations in cell structure, genome size, and complexity. Below is a detailed explanation of the key differences.
Location
- Prokaryotes: DNA replication in prokaryotes occurs in the cytoplasm because prokaryotes lack a membrane-bound nucleus. Their genetic material is free-floating in the cytoplasm, often in a region called the nucleoid.
- Eukaryotes: DNA replication in Eukaryotic cells takes place in the nucleus, where the DNA is compartmentalized. This nuclear localization ensures that replication is spatially regulated and protected from cytoplasmic processes.
Structure of DNA
Prokaryotes: Typically possess a single, circular chromosome that is relatively small. The DNA is associated with fewer proteins. Due to this, it is less compact and more accessible for replication machinery.
Eukaryotes: Have multiple linear chromosomes. For example, humans have 46 chromosomes. The DNA is tightly packaged into chromatin by wrapping around histone proteins, forming nucleosomes. This packaging complicates replication but protects the DNA and regulates gene expression.
Origin of Replication
Prokaryotes: Replication begins at a single origin of replication (called oriC in bacteria like E. coli). This single starting point is sufficient for their smaller, circular genomes.
Eukaryotes: They have multiple origins of replication per chromosome to accommodate their much larger genomes. Each origin initiates replication independently.
DNA Replication Meselson and Stahl Experiment
Matthew Meselson and Franklin Stahl experimented on DNA replication in 1958.
The experiment involved DNA replication in bacteria. They labeled E. coli bacteria with the heavy nitrogen isotope ¹⁵N. Then, they transferred them to a medium with light ¹⁴N. They observed the density of the resulting DNA via density gradient centrifugation.
The results showed intermediate-density DNA after one replication and both intermediate and light-density DNA after subsequent generations. It confirmed the semi-conservative model of DNA replication.
DNA Replication Errors
DNA replication errors are mistakes that occur when a DNA polymerase makes a mistake. They also occur when cellular processes like strand slippage cause issues.
DNA polymerases have proofreading and mismatch repair (MMR) systems to correct these errors. But failures in these processes can lead to mutations. They can cause permanent changes to the DNA sequence.
Types of DNA Replication Errors
There are several types of DNA errors that can occur during replication.
Base Pair Mismatches
The wrong base is paired with the template strand. For example, adenine pairs with cytosine instead of thymine.
Insertions
Sometimes, extra nucleotide bases are mistakenly added to the newly synthesized strand, which causes a replication error.
Deletions
When nucleotide bases are omitted or skipped during the synthesis of the new strand, it causes an error in the replication of DNA.
Consequences of Uncorrected Errors
- Mutation: If a mismatch is not corrected by the repair systems, it becomes a permanent mutation upon subsequent replication.
- Cancer: Replication errors, particularly those that lead to chromosomal rearrangements or activation of cancer-promoting genes, are a significant factor in the development of cancer.
- Genetic Diseases: Errors in germline cells can lead to heritable genetic diseases.
Conclusion
DNA replication is the semi-conservative biological process where a double-stranded DNA molecule is duplicated into two identical molecules. This process ensures each new cell receives a complete and accurate copy of the genetic information. The concept map helps you understand the process in easy
Prepare for Biology Exam With Expert Tutors
Are you struggling with biology and stressed about how to prepare for exams? We can help you. We have expert online science tutors to help you learn biology easily. You will get targeted preparation for your exams.
