DNA Replication in Prokaryotes
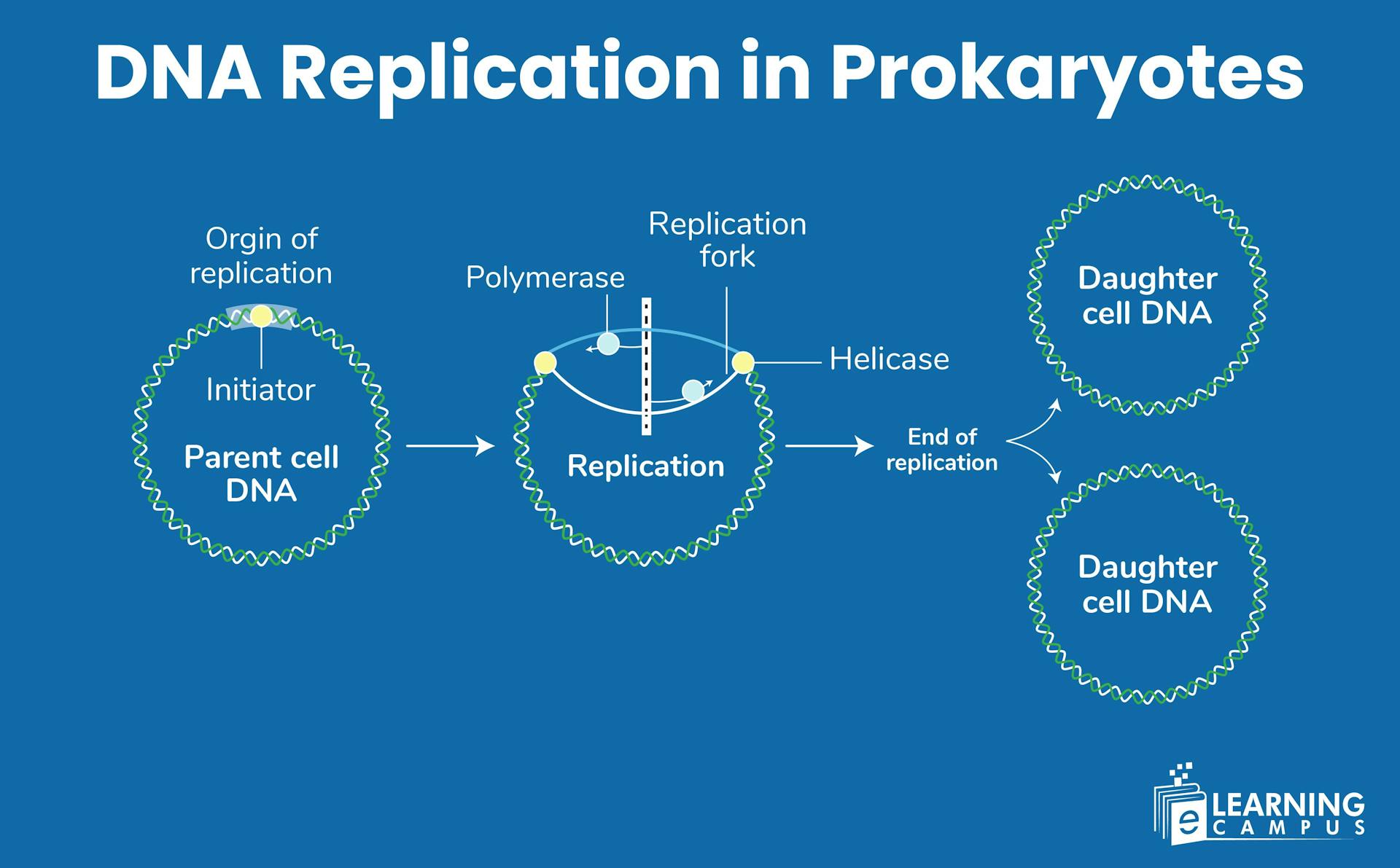
Have you ever thought about how microscopic prokaryotes, which are smaller than a speck of dust, flawlessly copy their entire genetic code in just minutes? DNA replication in Prokaryotes is not just a cellular task, but it is a molecular mystery that is carried out by enzymes and proteins working in coordination.
If you are curious about the transmission of genetic material in prokaryotes, this blog is for you. In this blog, we will explore how microorganism like bacteria replicate their DNA.
DNA Replication Process in Prokaryotes
DNA replication in prokaryotes is divided into three main phases: Initiation, Elongation, and Termination. Each phase has specific steps and enzymes involved.
Initiation
Initiation is the first process in prokaryotic DNA replication. The cell prepares to begin copying its DNA by identifying where to start and opening up the DNA structure.
Key enzymes in Initiation:
- DnaA: Initiator protein that recognizes oriC
- DnaB: Main helicase enzyme
- DnaC: Helicase loader
- SSB proteins: Single-strand binding proteins
- Primase (DnaG): RNA primer synthesis
Recognition of Origin (oriC)
The bacterial chromosome has one specific starting point called oriC (origin of replication). The replication starts from here. DnaA proteins recognize and bind to specific sequences at oriC. Multiple DnaA proteins accumulate at this site, forming a complex.
DNA Unwinding Begins
DnaA proteins cause the DNA to bend and partially unwind. The AT-rich regions (weaker bonds) at oriC begin to separate first. This creates an initial bubble of separated DNA strands.
Helicase Loading
DnaB helicase (the main unwinding enzyme) is loaded onto each separated strand. DnaC proteins help load DnaB helicase onto the DNA. Two helicases move in opposite directions from the origin.
Replication Fork Formation
As helicases continue unwinding, they create replication forks. Single-strand binding proteins (SSB) immediately coat the exposed single strands. SSB proteins prevent the strands from re-forming base pairs or forming secondary structures
Primer Synthesis
Primase (DnaG) synthesizes short RNA primers (8-12 nucleotides long). These primers are essential because DNA polymerase cannot start synthesis without a 3'-OH group. Primers are added to both template strands at the replication fork.
In case you are struggling with science subjects, get help from expert online science tutors.
ELONGATION
The second phase in DNA replication of Prokaryotes is elongation. The DNA polymerases add nucleotides to create new DNA strands, but the process is different for each strand due to the directional nature of DNA synthesis.
Key enzymes in Elongation:
- DNA Polymerase III: Main replicative enzyme
- Sliding clamp (β-clamp): Processivity factor
- Clamp loader: Loads the sliding clamp onto the DNA
- Primase: Continues to make primers for the lagging strand
DNA Polymerase III Recruitment
DNA polymerase III holoenzyme is the main replicative enzyme. It can only add nucleotides in the 5' to 3' direction. The enzyme has 3' to 5' exonuclease activity for proofreading.
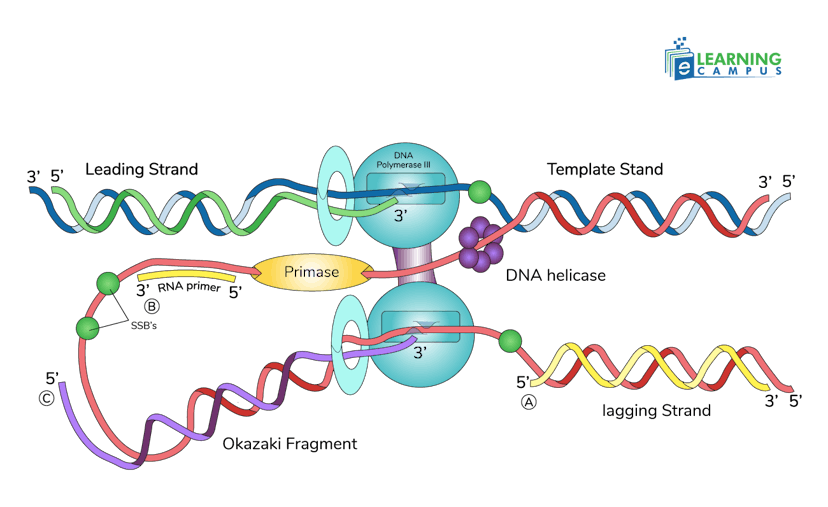
Leading Strand Synthesis
Continuous synthesis in the 5' to 3' direction. One primer is sufficient to start synthesis. DNA Pol III moves smoothly along the template. Synthesis occurs continuously as the helicase opens the DNA.
Lagging Strand Synthesis
Discontinuous synthesis due to the antiparallel nature of DNA. Must be synthesized in short fragments called Okazaki fragments. Each fragment is 1000-2000 nucleotides long in prokaryotes.
Process for each fragment
- Primase synthesizes a new RNA primer
- DNA Pol III synthesizes DNA from the primer
- Synthesis stops when it reaches the previous fragment
Simultaneous Synthesis
Both strands are synthesized simultaneously at the replication fork. The replisome (replication complex) coordinates both leading and lagging strand synthesis. The sliding clamp (β-clamp) keeps DNA Pol III attached to the DNA for processivity.
Proofreading
DNA Pol III has built-in 3' to 5' exonuclease activity. Incorrect nucleotides are immediately removed and replaced. This ensures high fidelity (accuracy) of replication
TERMINATION
Termination is the last stage of replication. The replication process is properly completed, with all RNA primers removed and DNA strands properly sealed.
Fork Collision
The two replication forks that started at oriC eventually meet. This occurs at the terminus region (ter) on the opposite side of the chromosome. Tus protein binds to ter sequences and acts as a fork trap. Tus protein allows forks to enter the terminus but prevents them from leaving prematurely.
Primer Removal
DNA Polymerase I removes each RNA primer and simultaneously fills the gap with DNA. This process is called nick translation.
Okazaki Fragment Joining
After primer removal, there are still breaks (nicks) in the DNA backbone. DNA ligase seals these nicks by forming phosphodiester bonds. This creates continuous DNA strands.
Chromosome Resolution
Sometimes the two circular chromosomes become interlinked (concatenated). Topoisomerases (like gyrase) help resolve these topological problems. XerC and XerD recombinases can separate concatenated chromosomes at specific sites.
Final Product
Two identical, circular DNA molecules form. Each contains one original strand and one newly synthesized strand (semiconservative replication). DNA is ready for distribution to daughter cells during cell division.
Characteristics of Prokaryotic DNA Replication
Speed and efficiency
The prokaryotic DNA replicates rapidly with efficiency. For example, E. coli replicates its 4.6 million base pairs in about 42 minutes. Each replication fork moves at approximately 1,000 nucleotides per second.
Accuracy
DNA polymerase III has proofreading capabilities (3' to 5' exonuclease activity), allowing it to remove mismatched nucleotides and ensure high fidelity.
Bidirectional replication
Starting from a single origin, two replication forks move in opposite directions, meeting at the terminus of the chromosome, where replication is completed.
Coordination
The replisome ensures that all components, such as helicase, primase, and polymerases, work together smoothly at the replication fork.
Why is DNA Replication Necessary in Prokaryotes
Prokaryotic cell DNA replication is essential for the successful transmission of genetic traits. It is the foundation for the continuation and propagation of the species. Without it, the genetic information would be lost or divided incorrectly between the new cells. It has importance in many ways.

Cell Division
Prokaryotes reproduce asexually through binary fission. This is a process where one cell divides into two identical daughter cells. For this to be successful, each new cell must have its own complete set of DNA. Therefore, a successful replication is crucial.
Genetic Continuity
DNA Replication ensures that the genetic material of the parent cell is accurately passed on to the daughter cells. It maintains the species' characteristics in prokaryotes.
No Genetic Information Loss
If DNA replication did not occur before cell division, the genetic information would be split and only partially passed on. It may prevent the daughter cells from functioning correctly. So, the prokaryotes' DNA replication is necessary.
Basis for Life
DNA carries the instructions for all cellular functions and life processes. Its Replication makes it possible to pass these essential instructions to new generations of cells and ensures life continues.
Conservation Across Species
The fundamental process and importance of DNA replication are conserved across all types of organisms, from single-celled prokaryotes to more complex life forms.
Proteins and Enzymes Involved in DNA Replication in Prokaryotes
Enzymes are protein molecules that speed up chemical reactions. They play an important role in the replication of DNA in Prokaryotes. Here are the enzymes that take part in the process.
Helicase
The helicase enzyme unzips the DNA double helix by breaking the hydrogen bonds between base pairs at the replication fork.
DNA Gyrase/Topoisomerase
This enzyme relieves the supercoiling and torsional stress created by unwinding the DNA helix, allowing replication to continue.
Primase
Primase synthesizes short RNA primers. It provides the free 3'-OH group that DNA polymerase needs to start synthesizing a new DNA strand.
DNA Polymerase III
DNA polymerase is the main enzyme responsible for synthesizing new DNA by adding nucleotides in the 5' to 3' direction.
DNA Polymerase I
DNA Polymerase I enzyme removes the RNA primers and replaces them with DNA. It plays an important role in DNA repair.
DNA Ligase
DNA Ligase joins the Okazaki fragments (short DNA segments on the lagging strand) by forming phosphodiester bonds, creating a continuous DNA strand.
Single-Strand Binding Proteins (SSBs)
Single-strand binding Proteins bind to the separated single strands of DNA. They prevent them from reannealing and protect them from degradation.
Sliding Clamp
Sliding Clamp is a ring-shaped protein that encircles the DNA and binds to DNA polymerase. It increases the stability and processivity, allowing it to synthesize long stretches of DNA.
Conclusion
DNA synthesis in prokaryotes, or replication, is a semi-conservative, semi-discontinuous process that starts at a single origin of replication (OriC) on a circular chromosome and proceeds bidirectionally. It completes in three steps: initiation, elongation, and termination.
Learn Science Online With Expert Tutors
Are you looking to hire an online science tutor for tests and exam preparation? You are at the right place. We have an expert science tutor with years of experience in helping students maximize their marks. You will get targeted preparation for your biology exams.