DNA Replication in Eukaryotes
Multicellular organisms accurately transfer their genetic material from parents to offspring. Ever wondered how a eukaryotic cell copies its massive genome with such precision? What happens when a cell cannot replicate its DNA correctly?
If you are confused about DNA replication in Eukaryotes, this guide can help clarify the process. In this blog, we will explain the mechanism of DNA replication in eukaryotes.
Stages of DNA Replication in Eukaryotes
DNA replication in eukaryotes involves three important stages. The three stages include initiation, elongation, and termination. Each stage comprises a complex process and action of enzymes.
Initiation
Initiation in the Eukaryotic cells is the process in which DNA replication starts. It includes the following steps.
Origin Recognition
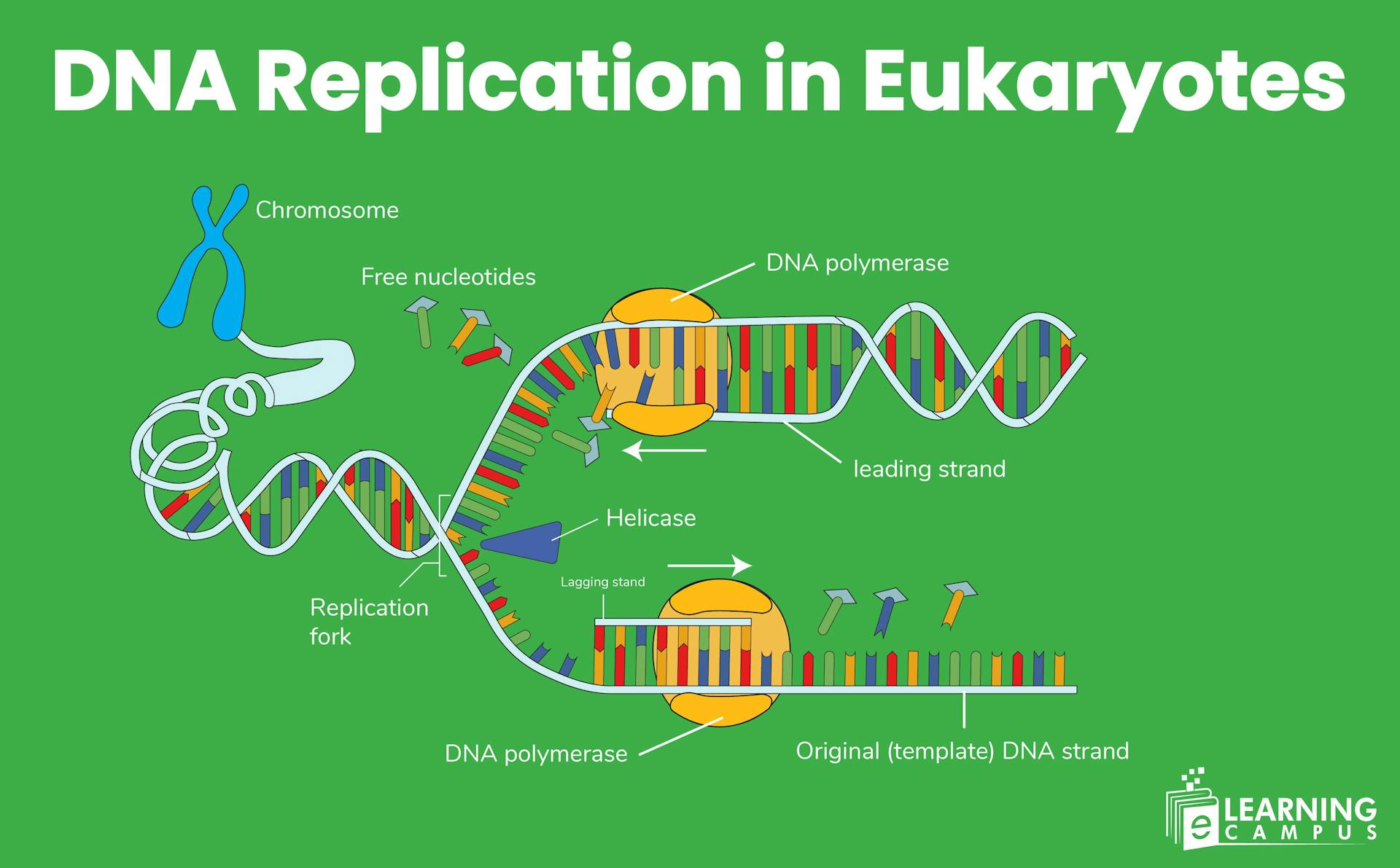
Replication begins at specific sites on the chromosome called origins of replication (ORs). These are specific sequences of DNA where the process of DNA replication begins. It allows unwinding of the double helix and the synthesis of new strands.
The origin of replication contains specific genetic elements called replicators. They are recognized by initiator proteins that recruit the machinery needed for DNA duplication.
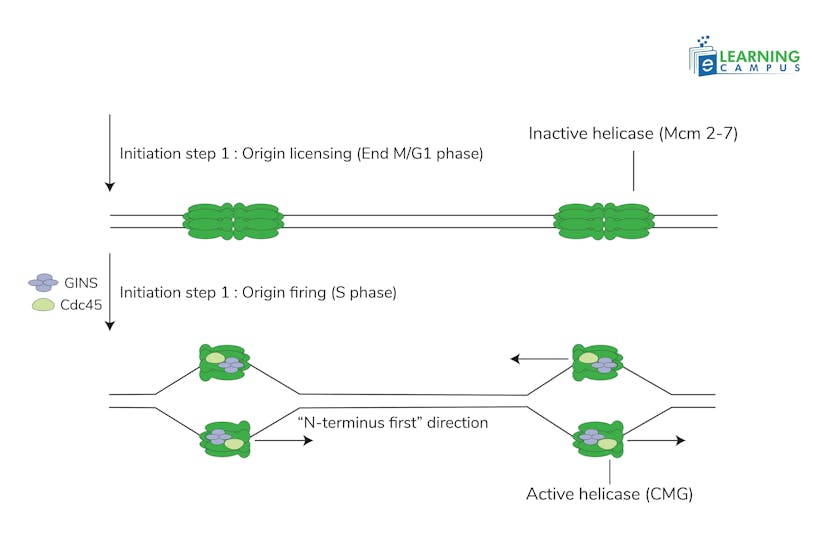
Pre-Replication Complex Assembly
In the G1 phase of the cell cycle, proteins like the Origin Recognition Complex (ORC), Cell Division Control Protein 6 (Cdc6), Chromatin licensing and DNA replication factor 1 (Cdt1), and the MCM helicase assemble at the OR to form the pre-replication complex.
Activation
In the S phase, kinases (a group of enzymes) like cyclin E/CDK2 activate the pre-replication complex by phosphorylating its components. It helps helicases to unwind the DNA.
Replication Bubble Formation
Helicases continue to unwind the DNA helix. Due to unwinding, a replication bubble with two replication forks is formed that moves in opposite directions. Single-strand binding proteins stabilize the separated strands.
Elongation
The second stage in eukaryotic DNA replication is elongation. In this stage, new DNA strands are synthesized by DNA polymerases, which add complementary nucleotides to the 3' end of a primer.
The process is facilitated by various proteins, such as helicases, single-strand binding proteins (RPA), and the sliding clamp PCNA (Proliferating Cell Nuclear Antigen). It includes the following steps.
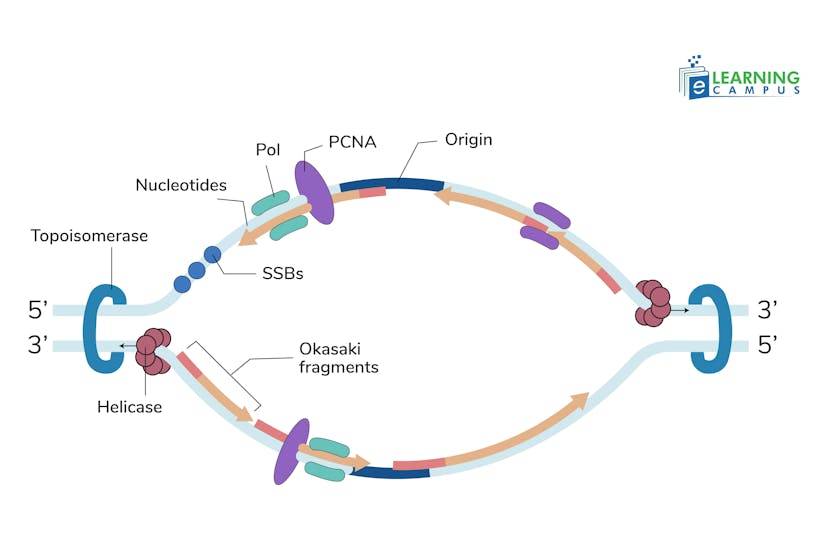
DNA Polymerase Alpha-Primase Complex
This complex initiates the process by synthesizing a short RNA primer on the single-stranded DNA template, followed by a short stretch of DNA.
Switch to Polymerase Delta
The polymerase alpha-primase complex then hands off the primer and DNA stretch to DNA polymerase delta (Pol δ) and the clamp protein PCNA, which is essential for stable, long-term synthesis.
Leading Strand Synthesis
The CMG helicase unwinds the DNA ahead of the replication fork. DNA polymerase moves along the leading strand, adding nucleotides continuously in the 5' to 3' direction.
Lagging Strand Synthesis
The lagging strand is synthesized discontinuously. Polymerase delta adds nucleotides to the 3' end and creates short Okazaki fragments.
RNA Primer Removal
Each Okazaki fragment is followed by a segment of DNA and an RNA primer. The RNA primer is then removed and replaced with DNA nucleotides by specialized enzymes.
DNA Ligase
Once the RNA primer is replaced, DNA ligase seals the final nick in the sugar-phosphate backbone, connecting the DNA fragments and completing the strand.
Termination
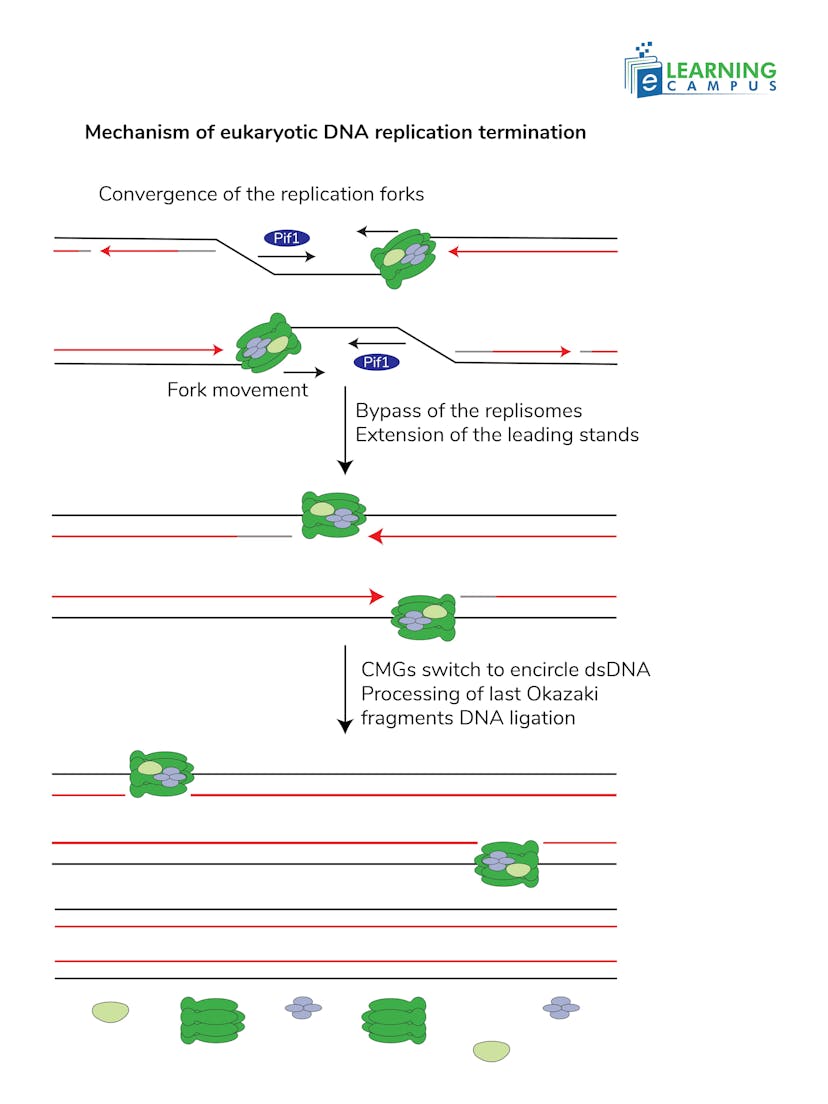
Replication Forks Meet
Replication terminates when the two replication forks moving from opposite directions meet in the middle of a replicon. There are multiple origins of replication, each with its own replication forks, that are active simultaneously.
Synthesis Completion
When the forks meet, the remaining gaps are filled in. The newly synthesized leading and lagging strands are bound together to create continuous sister chromatids.
Replisome Disassembly
The CMG helicase, a crucial part of the replisome, needs to be removed. This is facilitated by a dedicated pathway that involves ubiquitination and the action of the P97 protein (a segregase) to remove the helicase and other proteins from the DNA.
End Replication Problem
Eukaryotic chromosomes are linear, and the lagging strand cannot be fully replicated at the very end, creating the "end replication problem".
Telomerase Action
Telomerase, a specialized reverse transcriptase, extends the 3′ end of the lagging strand using an internal RNA template, thus preventing telomere shortening and maintaining chromosome integrity.
Decatenation
The process of replication creates entangled DNA molecules, or catenanes. A Type II topoisomerase resolves these constrictions. It ensures that the two newly formed DNA molecules can be separated.
Enzymes and Their Function in DNA Replication of Eukaryotes
In eukaryotic DNA replication, a complex set of enzymes works in a coordinated manner to duplicate the genome with high accuracy. Key enzymes carry out specific functions such as unwinding the DNA helix, synthesizing new DNA strands, removing RNA primers, and joining DNA fragments.
The following enzymes are involved in Eukaryotic DNA replication.
Helicase (Mcm2–7 complex)
The replicative helicase in eukaryotes is a ring-shaped protein complex called the Mini-Chromosome Maintenance (MCM) complex. It unwinds the DNA double helix by breaking the hydrogen bonds between the base pairs. It exposes the single-stranded DNA templates for replication.
Topoisomerase
This enzyme relieves the topological stress that occurs as the DNA double helix is unwound ahead of the replication fork. Two categories of topoisomerases are involved in this process.
- Topoisomerase I: This enzyme breaks a single strand and allows the DNA to turn to release tension.
- Topoisomerase II: It makes double-strand breaks to resolve DNA tangles and catenanes (interlocking DNA rings).
Primase
A primase enzyme is an RNA polymerase that synthesizes a short RNA primer to initiate DNA synthesis. The eukaryotic primase is part of the DNA Polymerase 𝛼-primase complex.
DNA Polymerase
DNA polymerase is a complex of proteins. This complex contains the primase, which synthesizes the RNA primer. After the primer is laid down, Pol α adds a short stretch of DNA nucleotides before handing off to the more processive polymerases.
Proliferating Cell Nuclear Antigen (PCNA)
This protein forms a ring-shaped sliding clamp that encircles the DNA and tethers the DNA polymerase to the template strand. It dramatically increases the processivity (the number of nucleotides added before detachment) of the polymerase to ensure efficient DNA synthesis.
Replication Factor C (RFC)
This is the clamp loader for PCNA. It recognizes the primer-template junction and uses ATP to load the PCNA ring onto the DNA.
RNase H
This enzyme specifically degrades the RNA portion of RNA-DNA hybrids, including the RNA primers from Okazaki fragments. RNase H2 recognizes and splits a single ribonucleotide embedded within a DNA duplex.
lap Endonuclease 1 (FEN1)
After RNase H removes most of the RNA primer, Polδ displaces the remaining RNA and DNA nucleotides by creating a flap. FEN1 removes this flap to prepare the nick for ligation.
DNA Ligase I
This enzyme seals the remaining nicks in the phosphodiester backbone by forming a covalent bond between the 3'-hydroxyl of one Okazaki fragment and the 5'-phosphate of the next. This is the final step in creating a continuous new DNA strand.
What Are the Characteristics of Eukaryotic DNA Replication
Multiple Origins
There are multiple origins of replication in Eukaryotes. Because eukaryotic genomes are large and linear. Due to this, the entire genome is duplicated once per cell division cycle.
Semi-Conservative
DNA replication in eukaryotes is semi-conservative. It means that each new DNA molecule consists of one original (parental) strand and one newly synthesized strand.
Bidirectional Replication
Eukaryotic DNA replication is bidirectional. From each origin, two replication forks form and move in opposite, bidirectional directions. They create a replication bubble.
S-Phase Specific
In eukaryotes, DNA replication occurs only during the S (synthesis) phase of the cell cycle. A pre-replication complex that forms earlier is activated only in the S phase to prevent re-replication.
Nucleus Location
Unlike the prokaryotic cell, DNA replication takes place in the nucleus of eukaryotic cells.
Linear Chromosomes and Telomeres
Eukaryotic chromosomes are linear, and a specialized enzyme called telomerase is required to add repetitive sequences to the ends . It prevents the loss of genetic information with each round of replication.
Complex Replisome
Eukaryotic replication involves a large, coordinated replisome complex of many proteins, including DNA helicases, DNA polymerases, primase, and others.
Multiple DNA Polymerases
Instead of a single type, eukaryotes use several distinct DNA polymerases for their roles, such as DNA polymerase δ for the lagging strand and DNA polymerase ε for the leading strand.
Conclusion
Eukaryotic DNA replication is a precise process occurring in the S phase of the cell cycle that duplicates the linear chromosomes. It involves three main stages, including initiation, elongation, and termination.
Hire Expert Science Tutors online
Are you looking to hire an online science tutor for your kids? You are at the right place. We have expert tutors to make science concepts easy and understandable for kids. You will get targeted exam preparation and instructions.
